About Us
Our research group focuses on developing new mass spectrometric capabilities for measurement of biomolecules and the clinical applications of proteomics, lipidomics, and metabolomics. We aim for identification of early stage disease biomarkers and systems biological understanding of the pathogenic mechanisms underlying human diseases.

Latest News
Welcome Dr. Vivek Sarohi joining our group! Vivek obtaied his PhD in Biosciences from the IIT Mandi.
Proteomic method development
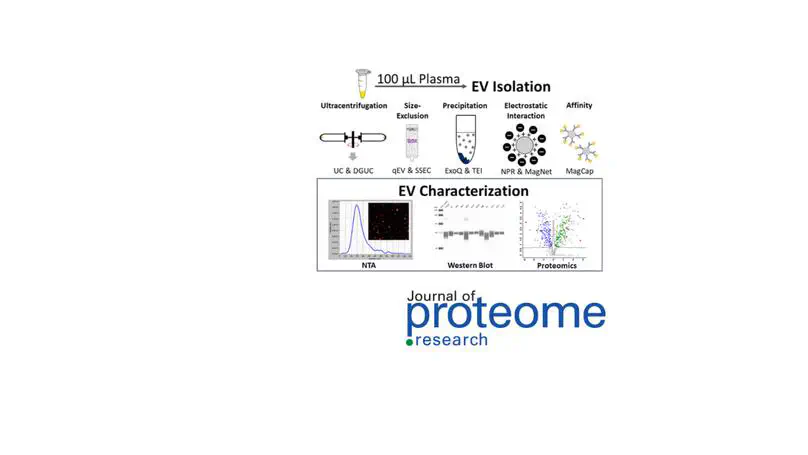
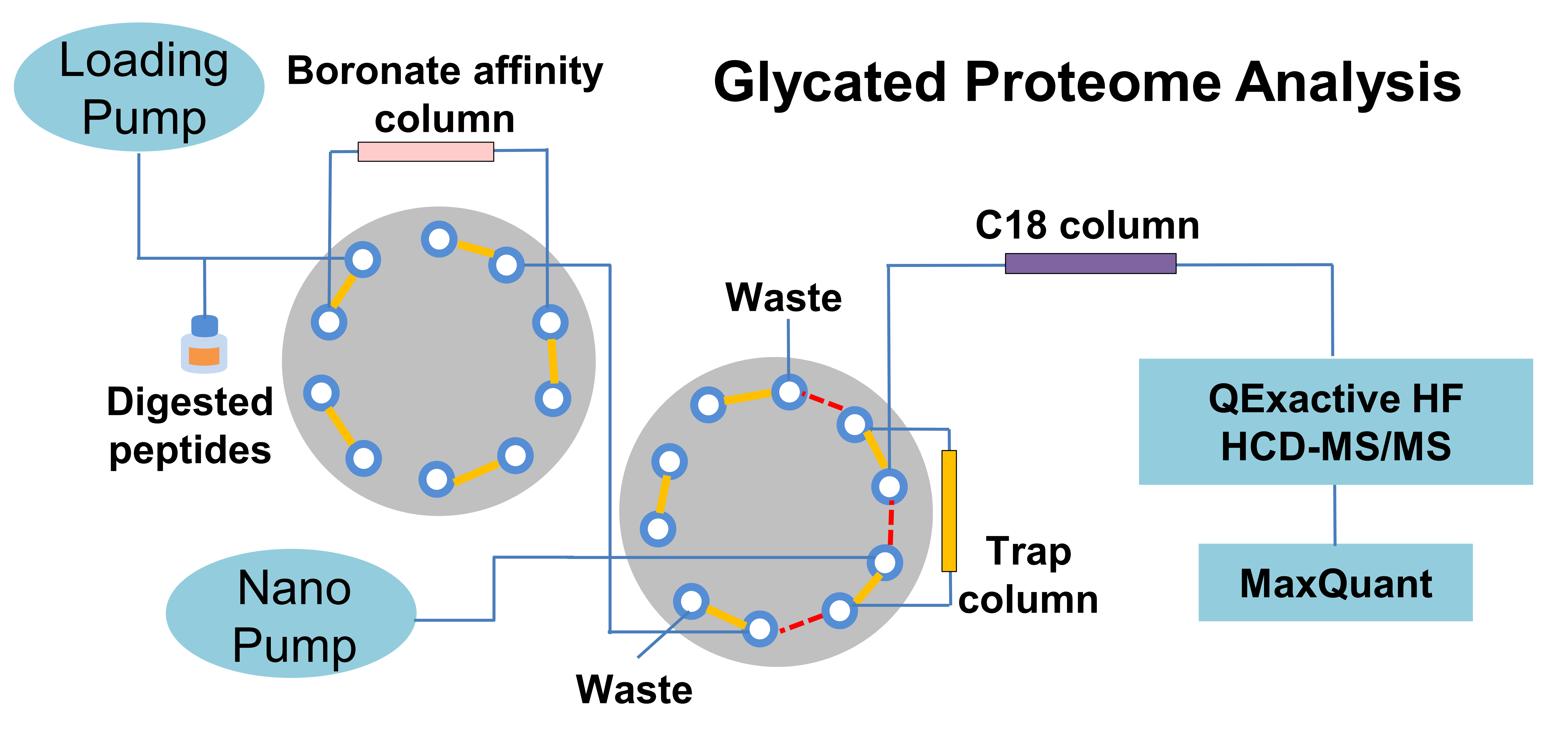
Analytical method development is driven by biomedical needs in measurement sensitivity, specificity and throughout. Many post-translational modifications to proteins are low abundant, accurate analysis of protein PTM requires specific enrichment methods. We developed online boronic affinity enrichment method for 2DLC-MS/MS analysis of glycated proteins, which has been used in identification of biomarkers to glycemic control and diabetic complications. We also developed laser capture microdissection-based methods for spatial proteomics to investigate the pathologies assoicated with specific cell types in a tissue. In addition, we developed highthroughout workflows for proteomic analysis of plasma and dried blood spot samples with improved proteome coverage, reproducibility and robustness. Continous method development is focused on proteomics for organelles, extracellular vesicles, and protein post-translational modifications.
Lipidomic method development
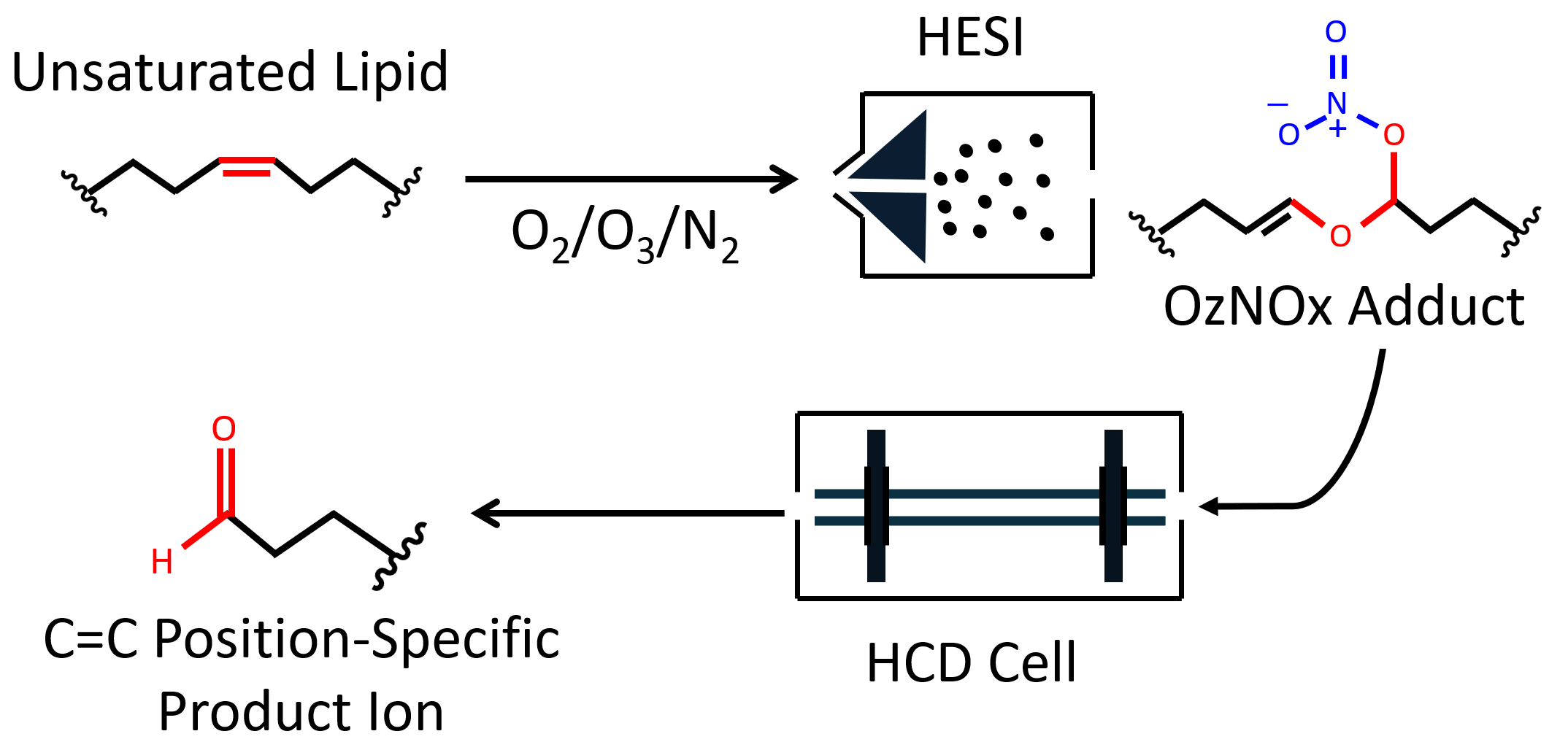
Lipids are structurally very diverse molecules. Different classes of lipids have different chemical properties, which makes their separation and identification from a complex biological sample very challenging. We developed methods for comprehensive lipidomic analysis, which include offline 2DLC separation coupled with high resolution mass spectrometry, novel ion chemistry OzNOxESI for determination of C=C position within unsaturated lipids, isobaric chemical labeling for more sensitive and multiplexed analysis of gangliosides, and very specific method for distinguishing isomeric oxylipins. Continued method development is focused on absolute quantification of lipids, lipidomics of tissue sections and extracellular vesicles, and modified lipids.
Biomarkers and pathogenic mechanisms for human diseases
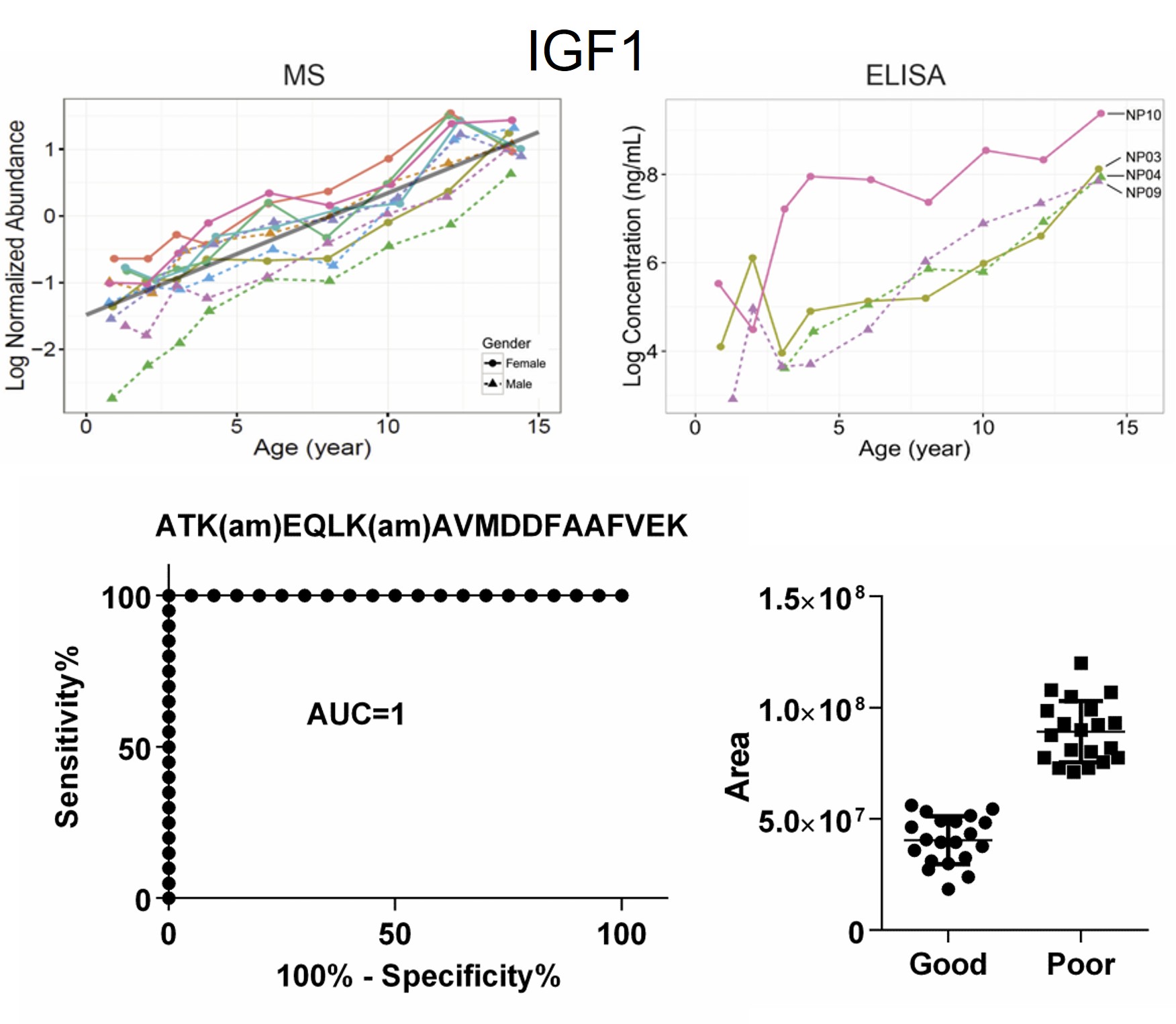
Reliable biomarkers are greatly needed in clinics for early diagnosis of human diseases. Our lab is the first that reported the most comprehensive profiling of longitudinal changes in plasma proteome during childhood development. Applying proteomics and lipidomics, we have identified panels of proteins that predict the onset of type 1 diabetes, diabetic complications and established gangliosides as a critical factor in hepatitus A virus entry into the host cells. Current research is focused on immunopeptides as neoantigens in type 1 diabetes, markers and mechanisms for tissue-specific diabetic complications and alcoholic heart diseases.
Molecular assessment of human performance
We developed novel omics marker panels for assessment of the immunity and inflammatory status. Through close collaboration with the Human Performance Lab of the Applacchian State University, we assessed the effect of consumption of various functional foods on boosting immunity and resolving inflammation induced by intensive exercises. Currently we are working with the Naval Health Research Center to evaluate the nutritional interventions to militrary training.

Informatics
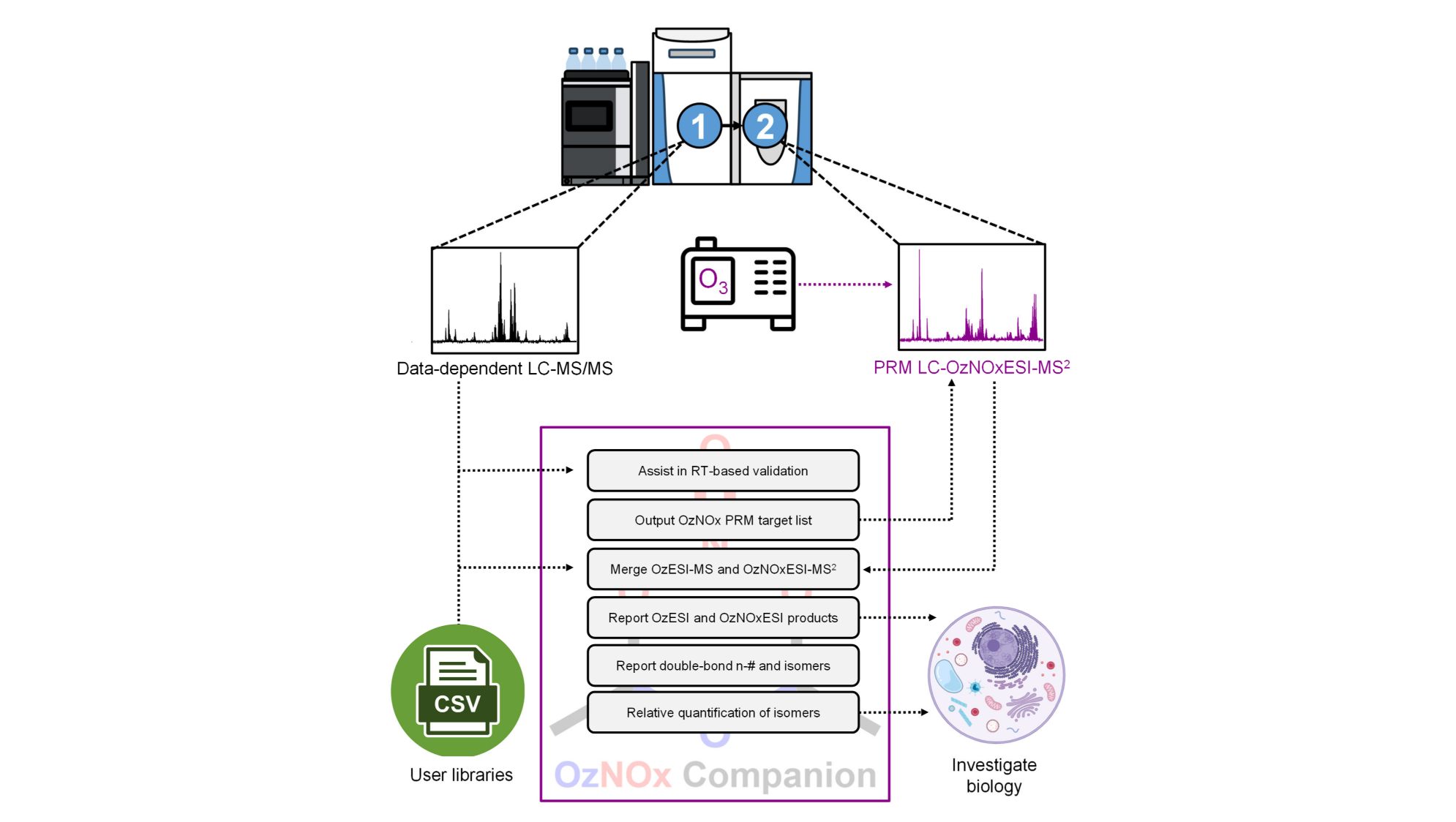
We developed software tools for automated processing of isotopic labeling mass spectrometry data and of the data generated by the novel OzNOxESI ion chemistry for C=C position analysis in lipids. Developing niche software to facilitate mass spectrometric data processing and data visualization will be a continous focus area of our research.
Join Us
Contact
Our research lab is in the UNCG Center for Translational Biomedical Research, located on the beautiful North Carolina Research Campus in the thriving downtown Kannapolis, a suburb of Charlotte.
- q_zhang2@uncg.edu
- 704-250-5803
- 600 Laureate Way, Kannapolis, NC 28081
- M-F 9:00 to 6:00